© Pint of Science, 2025. All rights reserved.
Since the 12th December 2019, when the first patient was hospitalised, it’s clear that coronavirus has spread worldwide – along with conspiracy theories and demand for vaccines and treatments.
“Coronavirus”, “covid-19” and “SARS-CoV-2” have all been used when reporting on this pandemic, but there are some nuances to what these words mean. The virus itself is called “SARS-CoV-2”, while the group of viruses SARS-CoV-2 is part of are termed “coronaviruses” – like humans are considered part of a larger group of primates. “Covid-19” is the name of the respiratory disease caused by SARS-CoV-2, much like MERS and SARS.
In just weeks, the scientific community started studying covid-19 and made substantial breakthroughs that are leading the world towards a better understanding of SARS-CoV-2, and in turn to potential treatments and vaccines. We now know that this virus is a coronavirus, made up of RNA (which holds genetic information like DNA does), a viral envelope (a double layer of greasy molecules called lipids) and several proteins embedded in the envelope. One of the major areas of study is the structural biology of the SARS-CoV-2 proteins.
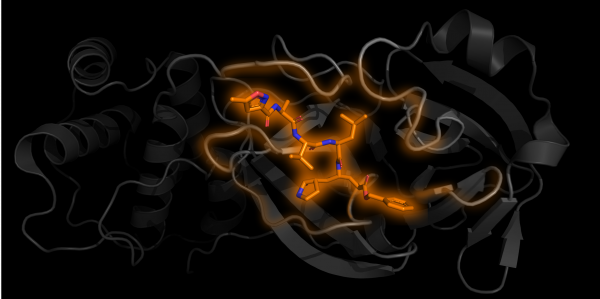
One subunit of Mpro in grey with an inhibitor N3 bound in orange PDB 6LU7
Structural biology is the study of how biological molecules arrange themselves and what they look like. Proteins are nanosized biological “tools” produced and used by cells and viruses. Their jobs are varied. For example, once inside the cell, the virus loses its lipid envelope and releases its genetic information in the form of RNA, which is converted into one long chain of proteins by the host cell machinery. For the virus to spread, this long protein chain needs to be chopped into separate parts to form more viral proteins. The protein used for the chopping is called Mpro, which acts as a pair of molecular scissors. First Mpro cuts itself free, then it can move on to cutting other parts of the chain.
Once scientists know what these proteins do and how they do it, they can work out which ones could be targeted by a medicine. Medicines and drugs typically work by changing or stopping protein doing its job. If a medicine could stop Mpro from cutting up that long chain of proteins, it would prevent the proteins getting free and the virus from spreading beyond that cell.
There are various approaches to drug development, but they mostly involve iterative cycles of (1) screening a library of chemicals for their activity (i.e. how well they change how the protein works), (2) identifying the most active chemicals, (3) attempting to optimise them by increasing their activity, “drug-like”-ness, safety or specificity to the protein of interest, (4) building up a new optimised library, and then again (1) screening for activity. For the optimisation step, it is highly beneficial for the drug developers to have structural data; that is to understand the shape and physical properties of the protein. Instead of adding different chemical groups somewhat at random, they can work more methodically – what shape chemical might fit best? By finding the structure of the protein with the chemical bound, the scientists can more easily “see” how the two are interacting.
As of the 29th of April 2020, 137 structures of SARS-CoV-2 proteins have been found, 121 of which are different structures of Mpro. Some of these structures have chemicals bound into the “active site” of the protein, the equivalent of the cutting edge of the scissor blades. Just like if the blades of actual scissors were blocked, these bound chemicals get in the way and prevent the Mpro segment from cutting the protein chain. This provides the starting point for potential treatments and future medicines, as scientists can work out where they bind, how they stop the protein from working and hopefully how they can be optimised.
Structural data can also allow scientists to perform some of the drug discovery steps computationally, which can speed up the process by reducing the amount of expensive and time-consuming experimental testing in a lab. This virtual screening allows chemists to quickly sift through huge libraries of compounds and focus on those predicted to have high activity or binding potential. One interesting virtual screening technique is “fragment screening”, which is being widely used to target Mpro. This is where small sub-parts of chemicals are added to the protein structure. If they are predicted to bind somewhere favourable, the small pieces can move from virtual to real-life testing and be built-up into bigger drug-like chemicals or provide ideas for the optimisation of other chemicals.
It’s important to note that it takes a long time and a lot of work to get from these chemical starting points, through safety testing and clinical trials towards a widely available medicine. Prior to this outbreak, there were already Mpro structures available, one of which, from 2003, is almost identical to the latest ones from SARS-CoV-2 (up to 96% similar) and the active site is 100% identical. Much like you might have several pairs of scissors at home – their handles might change, but the blades stay the same. These earlier structures were being used to develop medicines, but primarily financial barriers prevented this from being explored fully. Had there been more investment following the SARS and MERS epidemics, we might have been a lot closer to having a treatment ready for this one. What other potentially vital medicines could we be missing out on due to barriers and lack of investment?
Despite this delay in resources, the progress we’re seeing now by identifying these structures and chemicals represents the early stages of a hopefully successful journey to effective coronavirus treatments.
About the Author
Alex Holmes (she/her) is a PhD student at the Faculty of Biological Sciences at the University of Leeds and volunteers as a Pint of Science team leader for the 'Our Body' themed events in Leeds. She’s found herself working somewhere between biology, chemistry and computational science to research the structure and function of a family proteins. Twitter: @aomholmes


